A new study from the Université de Montréal and collaborators has uncovered an unexpected diversity in how even closely related bacterial species elongate their cells, a fundamental process for their growth and survival. This discovery has significant implications for understanding bacterial evolution, adaptation, and could pave the way for new antibiotic strategies.
Context
Bacteria exhibit a remarkable array of shapes and sizes, crucial for their different lifestyles and environments. A key process determining these shapes is cell elongation – how bacteria grow longer before they divide. For decades, scientists believed that the mechanisms driving cell elongation were largely conserved, especially among species that are close relatives on the evolutionary tree. Most of our understanding came from studying a few “model” bacterial species, leading to assumptions that others would behave similarly.
The Problem to Solve
The research team, led by scientists at the Université de Montréal and the Institut Courtois d’innovation biomédicale, questioned this long-held assumption. They aimed to investigate the diversity of cell elongation mechanisms in a group of closely related bacteria, the Caulobacteraceae family. The central question was: do these bacterial cousins all grow in the same way, or is there more variation – or “phenotypic plasticity” – than previously thought? Understanding this could reveal how bacteria adapt their growth strategies to different conditions and evolutionary pressures.
Experiments: A Glimpse into Bacterial Construction Sites
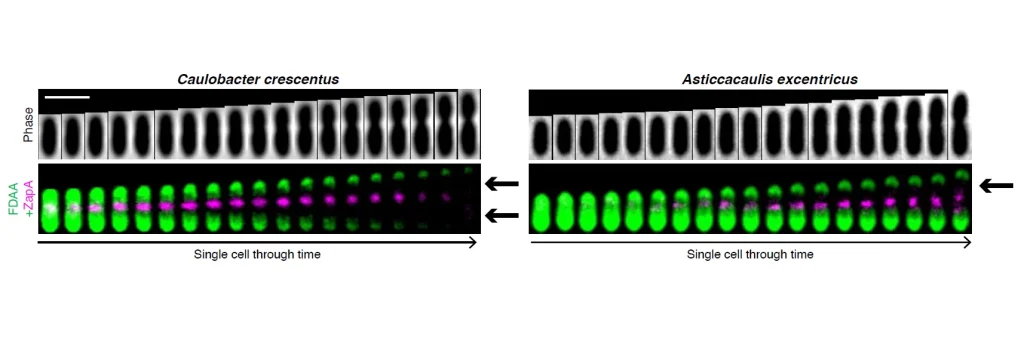
To watch these tiny organisms grow, the researchers, spearheaded by first author Dr. Marie Delaby, a postdoctoral fellow at the University of Montreal, employed sophisticated imaging techniques using fluorescent D-amino acids (FDAAs). These FDAAs get incorporated into the bacterial cell wall wherever new cell wall material is being actively synthesized. By tracking where these fluorescent signals appeared over time, the scientists could map out the “construction sites” for cell elongation in different bacterial species.
They combined these cell imaging techniques with genetic analysis and phylogenetic (evolutionary relationship) studies. This multi-pronged approach allowed them to not only see how different species elongated but also to understand the genetic underpinnings and evolutionary context of these differences.
Instead of a uniform method of elongation, even these closely related bacteria showed a startling variety of strategies. Some elongated from the middle outward in both directions, others only toward one end, and some began growing from one tip of the cell before switching to growth from midcell. These differences in where and how cells elongate highlight how flexible and diverse bacterial growth can be. They also investigated the localization of PBP2, a key protein involved in synthesizing the cell wall, and found that its position within the cell changed in ways that correlated with the different elongation modes. This suggested that the machinery responsible for cell growth is highly adaptable.
Conclusion: A New Appreciation of Bacterial Individuality
“This research might change our understanding of bacterial cell growth,” says Dr. Yves Brun, senior author of the study and Canada 150 Research Chair in Bacterial Cell Biology at the Université de Montréal and CI2B investigator. “We’ve shown that there’s a remarkable degree of phenotypic plasticity in how bacteria elongate, even among species we thought would be very similar. It’s clear that bacteria are much more flexible and diverse in their basic biological processes than previously appreciated.”
Relevance to Human Health
The mechanisms of bacterial cell wall synthesis and elongation are prime targets of some of our most effective antibiotics, such as penicillin. Discovering such unexpected diversity and plasticity in these fundamental processes has crucial implications for human health.
“If different bacteria build their cell walls in varied ways, it means that antibiotics targeting these processes might have different levels of effectiveness against them,” explains Dr. Delaby. “Understanding this diversity is critical for developing new antibiotics and combating antibiotic resistance. By learning about these alternative growth strategies, we might be able to identify new, more specific targets for future drugs, or predict how bacteria become resistant.”
The study is a collaboration with Francisco Pulido and Dr. Frederic J Veyrier from the Institut National de la Recherche Scientifique (INRS) and Dr. Michael S. VanNieuwenhze from Indiana University.
The full study is available in open access on the Nature Communications website.

Professor Yves Brun, PhD

Marie Delaby, PhD. Postdoctoral Researcher

